BART
Binding Analysis for Regulation of Transcription
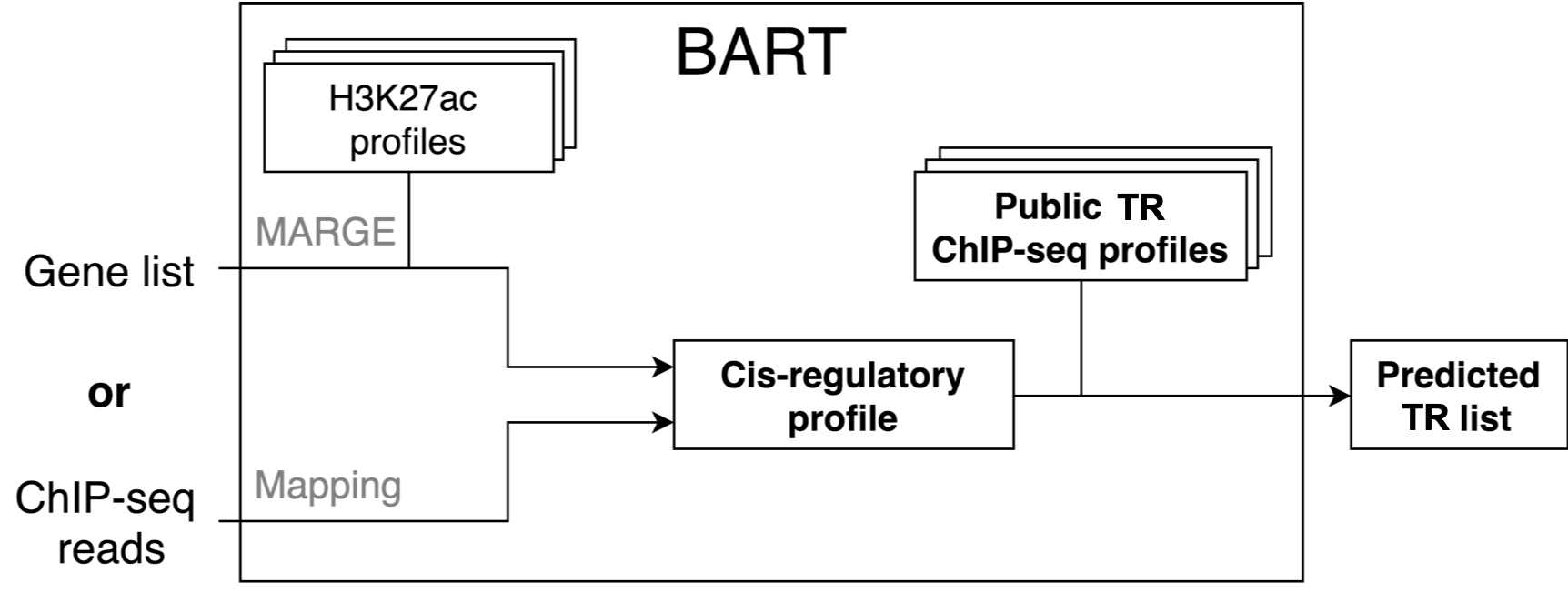
This web server is to perform BART, a bioinformatics tool for predicting functional factors (including transcription factors and chromatin regulators) that bind at cis-regulatory regions to regulate gene expression in human or mouse, taking a query gene list or a ChIP-seq dataset as input.
The current version is BART 2.0.
BART 2.0 uses over 7,000 human ChIP-seq datasets and over 5,000 mouse ChIP-seq datasets collected from the public domain to make the prediction. Updates in the current version include an optimized feature selection algorithm with Adaptive Lasso, resulting in much faster performance. BART is implemented in Python and distributed as an open-source package along with necessary data libraries. BART package is available on Github